
Recently, a research team from Institute of Health and Medical Technology, Hefei Institutes of Physical Science (HFIPS) of Chinese Academy of Sciences (CAS), developed an analytical service platform called CRISPRimmunity, which was an interactive web server for identifying important molecular events related to CRISPR and regulators of genome editing systems.
The research results were published in Nucleic Acids Research.
The new platform CRISPRimmunity was for the integrated analysis and prediction of CRISPR-Cas and Anti-CRISPR systems. It included customized databases with annotations for known Anti-CRISPR proteins, Anti-CRISPR-associated proteins, class II CRISPR-Cas systems, CRISPR array types, HTH structural domains and mobile genetic elements. These resources allowed for the molecular events in the co-evolution of CRISPR-Cas and Anti-CRISPR systems to be studied.
To improve prediction accuracy, the platform used strategies such as homology analysis, association analysis and self-targeting in prophage regions to predict Anti-CRISPR proteins. When tested on data from 99 experimentally validated Acrs and 676 non-Acrs, CRISPRimmunity achieved an accuracy of 0.997 for Anti-CRISPR protein prediction.
Furthermore, the platform provided the first ab initio prediction algorithm for class II CRISPR-Cas systems, which identified several previously unknown proteins. This included four Cas9s with different PAM structural domains, one smaller Cpf1, 61 C2c10s and three unclassified novel V-type Cas proteins. Some of these proteins had already been experimentally validated for activity in vitro.
CRISPRimmunity has many advantages, according to the team. It is an easy-to-use web server that provides a comprehensive analysis platform for CRISPR-related research. It accurately predicts Anti-CRISPR proteins and identifies novel class II CRISPR-Cas loci, offering an evolutionary perspective of the CRISPR-Cas and anti-CRISPR systems. Unlike other resources that focus on specific domains only, CRISPRimmunity fills the gap by providing methods to identify new class II CRISPR-Cas systems.
The user-friendly interface offers various visualization and customization options, with machine-readable results and tutorials for users of all levels. The NCBI database provides access to annotated data on 18,408 fully sequenced and 235 Acr-containing bacteria, as well as 208,209 human gut microorganisms, including information on CRISPR-associated key molecular events. Users can download or view this data to aid future experimental design and data analysis. Also for computational biologists, a stand-alone version of CRISPRimmunity is available on Github for bulk data mining.
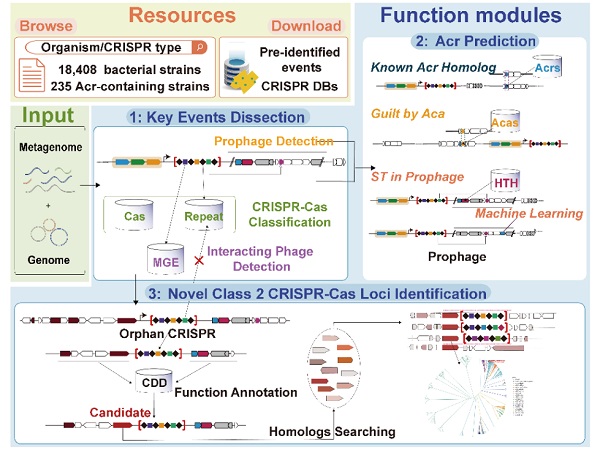
CRISPRimmunity modules and pipeline (Image by ZHANG Fan)