
Do you ever wonder how researchers identify bacterial infections? Traditionally, they collect samples from the infected site, grow the bacteria in a lab, and analyze them using a method called MALDI-ToF-MS. Although accurate, this method is time-consuming with a detection process that takes 1-3 days.
A research team from Hefei Institutes of Physical Science of Chinese Academy of Sciences, come up with a faster solution and published the way in Analyst as inside front cover.
They've developed a method using proton transfer reaction-mass spectrometry (PTR-MS) and fast gas chromatography-proton transfer reaction-mass spectrometry (FGC-PTR-MS) to detect volatile organic compounds (VOCs) emitted by 6 types of bacteria.
"We found each type of bacteria has its own unique smell," said Prof. SHEN Chengyin who led the team, "This can be used for rapid identification of bacterial species."
In this study, researchers focused on common pathogenic bacteria in patients with ventilator-associated pneumonia (VAP), a typical respiratory infection. The bacteria were rapidly detected using PTR-MS after cultivation, and characteristic ions were qualitatively analyzed using FGC-PTR-MS.
The experimental results indicate that after 3 hours of quantitative cultivation, the VOCs released by 6 bacteria can differentiate, and utilizing these differentiated VOCs enables significant differentiation of bacterial strains. This research demonstrates the feasibility of rapidly identifying bacterial strains through bacterial VOCs.
"We hope that the relevant technologies can help doctors formulate treatment plans promptly and enhance patient survival rates." said Dr. Xu Wei, first author of the paper.
The research was selected by Analyst as inside front cover. (Image by XU Wei)
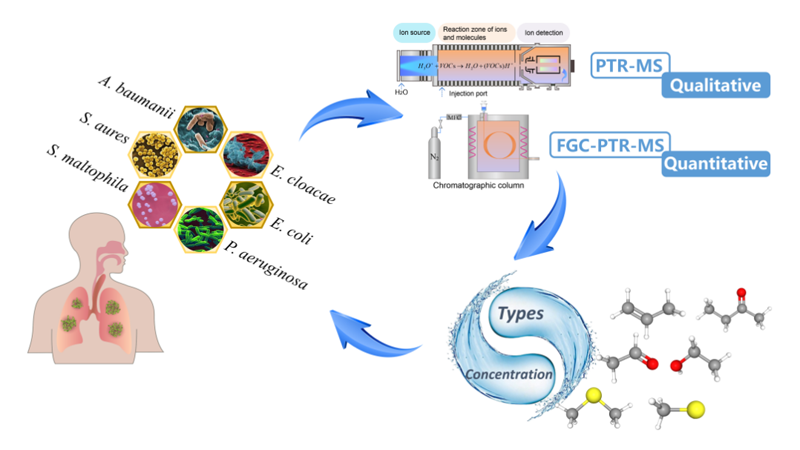
Schematic diagram of experimental process and results. (Image by XU Wei)